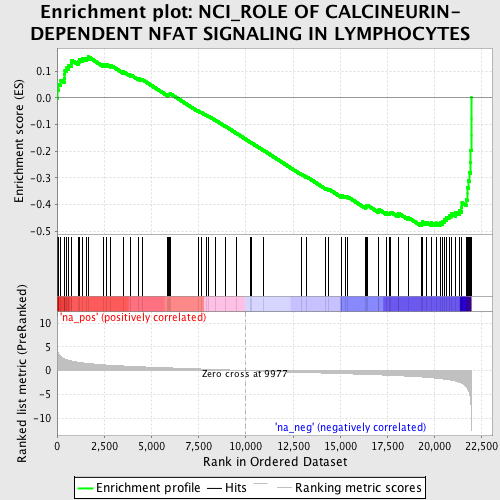
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | set02_ATM_minus_versus_BT_ATM_minus |
| Phenotype | NoPhenotypeAvailable |
| Upregulated in class | na_neg |
| GeneSet | NCI_ROLE OF CALCINEURIN-DEPENDENT NFAT SIGNALING IN LYMPHOCYTES |
| Enrichment Score (ES) | -0.47989893 |
| Normalized Enrichment Score (NES) | -1.9067202 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.033152204 |
| FWER p-Value | 0.335 |

| PROBE | GENE SYMBOL | GENE_TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|---|
| 1 | IL2RA | 15 | 4.747 | 0.0302 | No | ||
| 2 | NUP214 | 64 | 3.714 | 0.0522 | No | ||
| 3 | ITCH | 197 | 2.938 | 0.0653 | No | ||
| 4 | CSNK2A1 | 381 | 2.459 | 0.0729 | No | ||
| 5 | PRKCA | 397 | 2.430 | 0.0880 | No | ||
| 6 | CASP3 | 413 | 2.406 | 0.1030 | No | ||
| 7 | CREBBP | 519 | 2.250 | 0.1128 | No | ||
| 8 | YWHAZ | 622 | 2.135 | 0.1221 | No | ||
| 9 | IL5 | 751 | 2.009 | 0.1293 | No | ||
| 10 | GSK3B | 765 | 1.997 | 0.1417 | No | ||
| 11 | CTLA4 | 1130 | 1.742 | 0.1364 | No | ||
| 12 | MAPK8 | 1169 | 1.724 | 0.1459 | No | ||
| 13 | CAMK4 | 1351 | 1.628 | 0.1482 | No | ||
| 14 | YWHAE | 1550 | 1.540 | 0.1492 | No | ||
| 15 | CSNK1A1 | 1654 | 1.498 | 0.1542 | No | ||
| 16 | PTPRK | 2458 | 1.209 | 0.1253 | No | ||
| 17 | PRKCE | 2610 | 1.173 | 0.1261 | No | ||
| 18 | POU2F1 | 2833 | 1.121 | 0.1232 | No | ||
| 19 | RAN | 3501 | 0.968 | 0.0990 | No | ||
| 20 | PIM1 | 3908 | 0.888 | 0.0862 | No | ||
| 21 | KPNB1 | 4321 | 0.818 | 0.0727 | No | ||
| 22 | BAD | 4507 | 0.786 | 0.0694 | No | ||
| 23 | EP300 | 5873 | 0.578 | 0.0107 | No | ||
| 24 | FASLG | 5895 | 0.575 | 0.0135 | No | ||
| 25 | NFATC3 | 5945 | 0.567 | 0.0149 | No | ||
| 26 | IL4 | 5986 | 0.560 | 0.0168 | No | ||
| 27 | E2F1 | 7467 | 0.357 | -0.0486 | No | ||
| 28 | XPO1 | 7628 | 0.335 | -0.0537 | No | ||
| 29 | CDK4 | 7918 | 0.298 | -0.0650 | No | ||
| 30 | FKBP8 | 8018 | 0.285 | -0.0677 | No | ||
| 31 | PPARG | 8381 | 0.235 | -0.0827 | No | ||
| 32 | MAP3K1 | 8906 | 0.161 | -0.1056 | No | ||
| 33 | IL3 | 9524 | 0.071 | -0.1334 | No | ||
| 34 | CALM2 | 10266 | -0.045 | -0.1670 | No | ||
| 35 | EGR4 | 10291 | -0.048 | -0.1677 | No | ||
| 36 | AKAP5 | 10935 | -0.146 | -0.1962 | No | ||
| 37 | TBX21 | 12958 | -0.386 | -0.2862 | No | ||
| 38 | MAP3K8 | 13234 | -0.417 | -0.2960 | No | ||
| 39 | RNF128 | 14244 | -0.538 | -0.3387 | No | ||
| 40 | CSF2 | 14405 | -0.557 | -0.3423 | No | ||
| 41 | YWHAQ | 15062 | -0.634 | -0.3682 | No | ||
| 42 | CALM1 | 15082 | -0.637 | -0.3649 | No | ||
| 43 | IFNG | 15303 | -0.665 | -0.3707 | No | ||
| 44 | MAPK3 | 15401 | -0.677 | -0.3707 | No | ||
| 45 | FOSL1 | 16325 | -0.800 | -0.4077 | No | ||
| 46 | CD40LG | 16375 | -0.807 | -0.4047 | No | ||
| 47 | YWHAB | 16464 | -0.818 | -0.4034 | No | ||
| 48 | PTGS2 | 17046 | -0.900 | -0.4241 | No | ||
| 49 | GATA3 | 17055 | -0.902 | -0.4186 | No | ||
| 50 | MAPK9 | 17448 | -0.962 | -0.4303 | No | ||
| 51 | PRKACA | 17631 | -0.996 | -0.4321 | No | ||
| 52 | YWHAH | 17687 | -1.006 | -0.4281 | No | ||
| 53 | DGKA | 18084 | -1.073 | -0.4392 | No | ||
| 54 | JUN | 18099 | -1.076 | -0.4328 | No | ||
| 55 | BCL2 | 18625 | -1.179 | -0.4492 | No | ||
| 56 | PRKCQ | 19298 | -1.339 | -0.4712 | Yes | ||
| 57 | TNF | 19341 | -1.351 | -0.4643 | Yes | ||
| 58 | CALM3 | 19591 | -1.421 | -0.4665 | Yes | ||
| 59 | FKBP1A | 19825 | -1.506 | -0.4673 | Yes | ||
| 60 | SFN | 20083 | -1.603 | -0.4686 | Yes | ||
| 61 | PRKCZ | 20308 | -1.703 | -0.4678 | Yes | ||
| 62 | SLC3A2 | 20423 | -1.750 | -0.4616 | Yes | ||
| 63 | PRKCD | 20514 | -1.805 | -0.4540 | Yes | ||
| 64 | MAF | 20636 | -1.867 | -0.4474 | Yes | ||
| 65 | PTPN1 | 20769 | -1.947 | -0.4407 | Yes | ||
| 66 | MEF2D | 20891 | -2.028 | -0.4331 | Yes | ||
| 67 | MAPK14 | 21120 | -2.209 | -0.4291 | Yes | ||
| 68 | FOXP3 | 21314 | -2.435 | -0.4221 | Yes | ||
| 69 | JUNB | 21440 | -2.670 | -0.4104 | Yes | ||
| 70 | CBLB | 21453 | -2.694 | -0.3934 | Yes | ||
| 71 | YWHAG | 21678 | -3.403 | -0.3815 | Yes | ||
| 72 | PRKCH | 21736 | -3.724 | -0.3599 | Yes | ||
| 73 | NFATC2 | 21737 | -3.736 | -0.3356 | Yes | ||
| 74 | IRF4 | 21795 | -4.216 | -0.3108 | Yes | ||
| 75 | NFATC1 | 21871 | -5.174 | -0.2805 | Yes | ||
| 76 | FOS | 21917 | -6.478 | -0.2404 | Yes | ||
| 77 | NR4A1 | 21923 | -6.633 | -0.1975 | Yes | ||
| 78 | EGR1 | 21942 | -8.847 | -0.1407 | Yes | ||
| 79 | EGR3 | 21943 | -9.744 | -0.0773 | Yes | ||
| 80 | EGR2 | 21944 | -11.902 | 0.0001 | Yes |
